Central German Meeting on Bioinformatics 2015
Institute of Computer Science, Halle (Saale), August 26-27, 2015
Bioinformatics groups in the region of central Germany, and especially in the three cities of Halle, Jena, and Leipzig, cover a broad spectrum of topics in bioinformatics research. This meeting shall bring together scientists from central Germany, foster exchange between our groups, and provide an opportunity to stimulate ongoing and to plan future collaborations. The Central German Meeting on Bioinformatics 2015 has its origins in workshops of the Jena Centre for Bioinformatics and is the first meeting in a series of annual meetings organized at Halle, Jena, and Leipzig. The meeting language is English.
We would like to invite abstracts for talks and posters on advances and challenges of current bioinformatics research. The meeting is open to contributions from all areas of bioinformatics. Abstracts should be at most one page long including references, should use the provided template, and should be submitted via easychair.
Meeting Leaflet and Abstract Book
Leaflet with program and list of posters (printed version provided at registration desk)
leaflet.pdf
(externe Datei)
Abstract book with list of keynotes and abstracts of invited talks, contributed talks and posters
abstract_book.pdf
(externe Datei)
Organizing institutions
Supporting Institutions & Sponsors
Registration
Participation in CGMB 2015 is free of charge.
However, we would kindly ask you to register for the meeting to give us an estimate of the number of participants we can expect.
Online registration is closed, but it will still be possible to register on-site.
Submission
When preparing your abstract, please use the following templates:
Using one of these templates, the length of your abstract (including references) must not exceed one page. Submission is PDF-only via EasyChair. If you use the Word template, please convert the final abstract to PDF format before submitting it.
When submitting an abstract, please indicate your favored type of presentation (talk or poster). Authors of accepted talk abstracts will be invited by the program committee to make either an oral presentation or a poster presentation. By submitting an abstract, authors agree on publication of abstracts on the meeting website.
Submission is closed.
Program
26.08.2015
27.08.2015
Key Dates
| July 15, 2015 | Due date for submission of abstracts (submission closed) |
| July 24, 2015 | Author notification about oral or poster presentation |
| August 26-27, 2015 | Central German Meeting on Bioinformatics 2015 |
Program Committee
Sebastian Böcker, Jena
Andreas Gogol-Döring, Halle/Leipzig
Jan Grau, Halle
Ivo Grosse, Halle
Jörg Hackermüller, Leipzig
Alexander Hinneburg, Halle
Steve Hoffmann, Leipzig
Janet Kelso, Leipzig
Birgitta König-Ries, Jena
Manja Marz, Jena
Martin Middendorf, Leipzig
Steffen Neumann, Halle
Stefan Posch, Halle
Uwe Scholz, Gatersleben
Stefan Schuster, Jena
Peter Stadler, Leipzig
Wolf Zimmermann, Halle
Organizing Committee
Jan Grau
Ivo Grosse
Alexander Hinneburg
Steffen Neumann
Stefan Posch
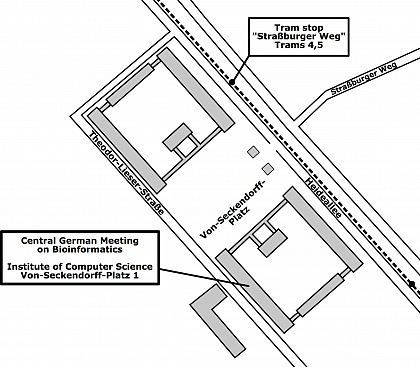
Venue
Martin Luther University Halle-Wittenberg
Institute of Computer Science
Lecture Hall 3.28
Von-Seckendorff-Platz 1
06120 Halle (Saale)
Germany
Contact
Kathleen Kletsch
Martin Luther University Halle-Wittenberg
Institute of Computer Science
Von-Seckendorff-Platz 1
06120 Halle (Saale)
Germany
Phone: ++49-345-55 24754
Fax: ++49-345-55 27039
E-Mail: cgmb@informatik.uni-halle.de