Research
Hochdurchsatzanalyse der Formeigenschaften von Pavement-Zellen
Das Ziel dieses Projektes ist die effiziente Analyse von Formmerkmalen von Zellen. In erster Linie schauen wir uns dabei Pavement-Zellen an, aber die entwickelte Methodik eignet sich prinzipiell für beliebige Zelltypen. Wir haben dazu ein neues Tool zur Hochdurchsatz-Analyse von Pavement-Zellen veröffentlicht, PaCeQuant, das Zellen in Mikroskopbildern automatisch segmentiert und automatisch eine Reihe von Formparametern extrahiert. Im Vergleich zu früher verfügbaren Tools erlaubt der integrierte Segmentierungsansatz dabei, auf einen zeitaufwändigen manuellen Annotationsschritt zu verzichten und größere, repräsentativere Datensätze als bislang üblich zu analysieren.
Publikationen:
- D. Mitra, S. Klemm, P. Kumari, J. Quegwer, B. Möller, Y. Poeschl, P. Pflug, G. Stamm, S. Abel, K. Bürstenbinder, ''Microtubule-associated protein IQ67 DOMAIN5 regulates morphogenesis of leaf pavement cells in Arabidopsis thaliana'', Journal of Experimental Botany, 70(2):529-543, January 2019.
- PaCeQuant: A Tool for High-Throughput Quantification of Pavement Cell Shape Characteristics ,
Kooperationspartner:
- Dr. Katharina Bürstenbinder , Department of Molecular Signal Processing, Leibniz Institute of Plant Biochemistry, Halle (Pressemitteilung des IPB )
- Dr. Yvonne Pöschl , Institute of Computer Science, Martin Luther University Halle-Wittenberg & German Integrative Research Center for Biodiversity (iDiv) Halle-Jena-Leipzig, Leipzig
PaCeQuant ist als Teil von MiToBo und damit als Erweiterung für ImageJ/Fiji implementiert.
- MiToBo website
- Homepage von PaCeQuant
Analyses und Verlaufsprognose von PET-Scans von Hodgkin Lymphome Patienten
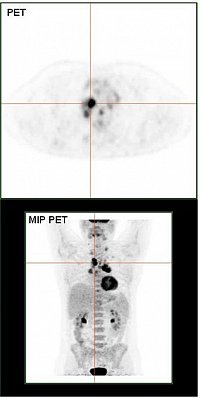
PET Scan (Quelle: https://commons.wikimedia.org/wiki/File:HodgkinLymphomPETCT.jpg, Modifiziert)
Das Hodgkin Symndrom ist eine maligner Tumor des Lymphsystems, dervorwiegend bei Kindern und Jugendlichen auftritt.Die Heilungsaussichten sind mittlerweise sehr gut, jedoch treten insbesonderaufgrund von Radiotherapie starke langfristige Nebenwirkung auf.In diesem Projekt soll auf der Basis von PET-Untersuchungen der Glukoseaufnahme (FDG-PET)prognostischer Faktoren identifiziert werden,die perspektivisch zur Risikobewertung und Therapiesteuerung genutzt werden sollen.Der entscheidene Ansatzpunkt für die Arbeiten ist der aktuelle Stand der Diagnosik,bei dem das Ausbereitungsmuster nur sehr grob berücksichtigt wird.
Das Ausbreitungsmuster des Tumor wird mittles automatischer Segmentierungsverfahrenund geeigneter Merkmale sowohl auf der Ebene von Körperregionen mit hoherGlukoseaufnahme (sowohl Normalgewebe wie auch Tumornester) und auf der Ebenedes gesamten PET scans der Patient*innen charakterisiert.Hierauf aufbauen werden Verfahren des machinellen Lernes genutzt, um Normal- vonTumorgewebe zu differenzieren und eine Verlaufsprognose automatisch zu bestimmen
Kooperationspartner:
Prof. Regine Kluge, Thomas GeorgiKlinik und Poliklinik für Nuklearmedizin,Universität Leipzig
Automatic segmentation of plant roots in minirhizotron images
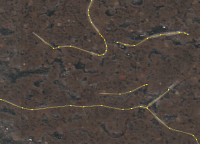
Minirhizotron image with segmentation results
Roots are an important organ of terrestrial plants as they provide vital functions. These include anchorage in the soil, resource uptake, -transport, and -storage. There is an intrinsical linkage to root production and mortality, accordingly root development is studied in biological research questions.
Minirhizotron systems are a non-destructive method of root measurement. They allow scientists to acquire images of roots directly in the soil and over time. Extracting the relevant data from the images, like root length and diameter, requires a precise root annotation. However, manual root annotation is a very time consuming and laborious task.
The aim of the project is to develop an automatic image segmentation procedure, which enables root detection in shorter time and with less (manual) effort. To achieve this we use different approaches like detection of relevant features, especially edges, and machine learning methods, e.g. deep learning. One big challenge is the inhomogenity of the soil as well as its water inclusions. Due to this the distinction between roots and soil gets more complicated even in manual annotation, while it makes an automatic root detection considerably more difficult.
The development of a reliable automatic root segmentation would enable the research community in biology to analyse more and bigger data sets in a resource efficent way and consequently helps to investigate biological relationships in plants to a greater extend.
Cooperations:
PD Dr. Alexandra Weigelt and Dr. Hongmei Chen,
Dept. of Systematic Botany and Functional Biodiversity,
Institute of Biology, Leipzig University
rhizoTrak: A new software tool for manual annotation of images from minirhizotrons

Minirhizotrons are a non-destructive way to monitor root growth. Transparent tubes are installed in the soil and a scanner takes images in these tubes. Due to its non-destructiveness, the system makes it easy to acquire time-series images which results in a large amount of image data. To analyse these data, researchers need to annotate the images manually which requires extensive effort.
Existing annotation software often has limited support for time-series annotation. Therefore, we develop the open source tool "rhizoTrak" for user-friendly, efficient and flexible manual annotation of complex time-series minirhizotron images. rhizoTrak builds on TrakEM2 while extending it with specific functionality for root image annotation. The tool is realized as a Fiji plugin. rhizoTrak relies on open data formats which cannot only be used for further analysis and statistical evaluations, but above all yields the perfect basis for developing and integrating further techniques, such as automatic segmentation.
Further details:
rhizoTrak website, https://prbio-hub.github.io/rhizoTrak/
Biological and bioinformatic analysis of TAL effectors in the Xanthomonas oryzae - rice interaction (DFG funded)

TAL effector

RVD binding specificities
PI: Dr. Jan Grau
This project aims at advancing our understanding of the interaction between plant pathogenic bacteria and their plant hosts. To this end, we will study transcription activator-like effector (TALE) function using the economically important model system Xanthomonas oryzae and rice (Oryza sativa). We follow an interdisciplinary approach combining biological and bioinformatics work packages. In the biological part we will identify TALEs from different, presently uncharacterized X. oryzae strains. Transcriptomics (RNA-seq) will reveal which target boxes of TALEs are functional in vivo and will also allow us to identify TALE target genes. Systematic reporter assays of target boxes in their promoter context will allow us to dissect different biological determinants of TALE function. The bioinformatic part will support TALE repertoire dissection and TALE target gene identification. In addition, the data generated in the biological part will be used in algorithms to further define in vivo TALE-DNA binding rules, and identify promoter characteristics or specific elements that are required for TALE-mediated gene-induction. Using this interlaced approach, the project will shed significant light on the biological activity of TALEs which will be instrumental to develop future resistant rice.
Collaborator:
PD Dr. Jens Boch, Dept. of Genetics, Institute of Biology, Martin Luther University Halle-Wittenberg
Detection and Tracking of Sub-cellular Structures in Microscope Images
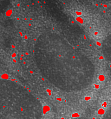
Microscope images of fluorescently labeled sub-cellular structures play an important role in biomedical research. They are of particular importance for understanding system processes within singular cells and their changes under varying environmental conditions. Relevant structures, like stress granules or p-bodies, show a large variation in their specific properties, e.g. in size or homogenity. Consequently techniques for automatic segmentation of these structures in microscope images likewise require a large flexibility and adaptivity. Amongst others we use adaptive algorithms based on wavelets to tackle this task. For tracking particles in videos they are combined with probabilistic filtering techniques.
Cooperations:
Section Molecular Cell Biology, Martin Luther University Halle-Wittenberg, Prof. Dr. Stefan Hüttelmaier
Segmentation of Cell Images with Active Contours
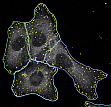
Single cells within large cell populations show a significant variety in their specific properties and behaviours, hence, large data sets need to be analyzed in biomedical research to cover the natural variation sufficiently well. Thereby in particular specifics of single cells are of high interest, and consequently qualitative as well as quantitative analysis steps, like the detection of sub-cellular structures, should always be done for individual cells. Indeed this requires the exact localization of single cells in populations and an exact segmentation of their contours. We solve this problem for a variety of different kinds of cells, amongst others also for neuronal cells, by adopting active contours (levelsets as well as snakes) and corresponding energy functionals.
Cooperations:
Section Molecular Cell Biology, Martin Luther University Halle-Wittenberg, Prof. Dr. Stefan Hüttelmaier
Alida and MiToBo

One important goal in our algorithm development is a large flexibility and also modularity to allow for easy reuse of developed components. At the same time implemented techniques are usually handed over to non-expert users immediately, hence, not only the algorithms by themselves, but likewise adequate user interfaces have to be provided. To this end we have developed a concept for an integrated development of functionality and user interfaces in data analysis named Alida (''Advanced Library for Integrated Development of Data Analysis Applications''). Alida defines a standard for implementing and executing data analysis operators, and by this supports amongst others the automatic generation of user interfaces and an automatic logging and documentation of analysis procedures.
Alida has been implemented in Java and prototypically in C++. The Java implementation serves as base for MiToBo, ''A Microscope Image Analysis Toolbox'', where our algorithms for microscope image analysis are collected. MiToBo subsumes operators for basic image processing like filters or morphological operators, and also more complex approaches like active contours or wavelets. In addition, specialized applications, e.g. for segmenting scratch assay images, are provided. MiToBo is based on ImageJ and fully compatible with the wide-spread software package.
More information: http://www.informatik.uni-halle.de/alida and http://www.informatik.uni-halle.de/mitobo

